- The "smart gene" hypothesis is an attempt to derive a model of gene flow which can accommodate many large-scale effects such as the evolution of sex, orthologous gene distribution, variation of mutation rates, concerted molecular drive, and the "punctuated" pattern of large-scale phenotypic change.
- However, the only model we can derive which both fully explains the effects and correlates with the observed patterns of gene flow, appears "stranger than we can suppose" to our normal perceptions of how events occur. Especially, the new model would imply that genes possess self-directed "drive", plus gene families can retain synchronism of sequence by 'action-at-distance' even after genes split into separate lines. More strangely, highly conserved genes are able to pass through modern day reproductive events as a form of probability distribution, unaffected by the events themselves.
- We can explain these strange effects as a form of intragenomic conflict between early evolving, highly conserved genes expressing the basic homologies of life and later evolving genes and DNA, which express highly variable details of life. The theory is that early evolving genes already achieve high sequence conservation, so they force later evolving genes and DNA to accept the burden of variability and change. Or this is the effect of phenotypic selection. Clearly, no genes act with foresight or intent.
Notes
:
- Critics find this theory very difficult to follow. Individuals struggle to be fit, and DNA is selected to express fitness. But change comes at an evolutionary cost. This cost is represented by a property of DNA to be "selfish" and maintain sequence in the face of change. Over the course of life, long established genes have found ways to resist change by shifting the burden of alteration to more recently evolved DNA. These are "smart" genes.
- The "smart gene" theory is incredibly controversial, but it still has nothing to do with "smart genes" in the sense of IQ, race, or intelligence. If anything, "smart genes" are very ancient, highly conserved genes, which evolved long before humans.
 I have added an essay form of this theory in 2.4 The Heuristic Process which people might find easier to follow.
I have added an essay form of this theory in 2.4 The Heuristic Process which people might find easier to follow.  I have added some math, which expresses the general idea. See Equations
I have added some math, which expresses the general idea. See Equations Or see the PDF file at model
Or see the PDF file at model
 ___________________
___________________
In this Section:
5.1 Gene Propagation examines two broad models of gene flow.
5.2 Gene "Selfishness" explains why sequences tend to self-perpetuate.
5.3 Gene Spectrum explains that the best "selfish" strategy for a gene would be to express a basic homology of life, essential in all individuals, that could not be selected out by single reproductive events.
5.4 Phase Shifts explains how we can show the total effects of gene flow in one model.
Return to
A New Model of Evolution Home Page

5.0 The "Smart Gene" Hypothesis
5.1 Gene Propagation
All current models of gene flow work on the simple assumptions that host fitness will reflect gene fitness.
a) If a gene produces an advantageous mutation, the host will prosper.
b) If it is a disruptive mutation, the host will die.
This model is very useful, except it requires that all genes pass or fail at a single reproductive event. Yet, many genes are homologous within species and orthologous across species. They exist in many individuals and will pass on anyway, just as 99% of human genes will pass on at each reproduction anyway. So, we need two models.
- Serial Propagation
: This is the intuitive model. It assumes that gene fitness is dependent on the fitness of the host phenotype in which the gene is ensconced. This model allows us to trace the fitness of a gene by the success of its host lineage to reproduce through many generations. Population genetics and computer simulations use this model exclusively.
- Radial Propagation:
This is a counterintuitive model. It assumes that gene fitness is independent of the fitness of any host lineage. This will occur once the gene expresses a homology essential to many individuals throughout populations, species, classes or even phyla. This model is used to study systematics or relatedness of species.
Fig 5.1a and 5.1b show the two types of gene propagation.
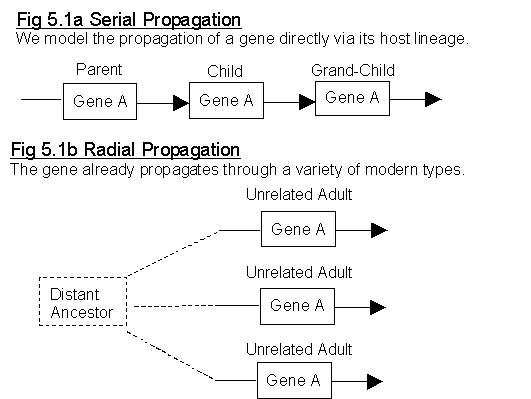
In nature, genes propagate by a combination of the two methods
- When genes first evolve, they are totally dependent on the lineage of their host phenotype to spread the new gene through a population. (Fig 5.1 a Serial Propagation. If the host lineage terminates, the gene lineage terminates.)
- Once genes "sweep" to fixation, they no longer depend on any individual lineage, or fitness of any one host, to propagate. (Fig 5.1 b Radial Propagation. Termination of a radial spoke will not bring "genetic death".)
Significantly, over the whole of life, most genes (those that survive) propagate radially, in that they do not depend on the fitness of any one individual or lineage for survival of the sequence. For example;
- The gene expressing the H4 protein is homologous to all eukaryotic life. Yet, it depends so little on the fitness of any host that this gene survived the K-T extinction unaltered. (If H4 is defective the host will die. But despite that many hosts die for many reasons, H4 as a sequence will avoid genetic death.)
- Each human has a unique DNA fingerprint, but of actual genes (3% of DNA) about 99% are common. Plus humans have found cultural and social ways to adapt. So, 99% of human genes radiate through many individuals with little alteration or likelihood of genetic death. (Unless human stupidity brings another global extinction.)
- Some 96% of genes in the ancestor great ape genome retained sequence while radiating into a variety of types including humans. Similarly, 70% of genes expressing mammal phylogeny radiate throughout a huge variety of types such that nothing short of global extinction could wipe them out.
- Core gene sets expressing deeper homologies, such as blood or photosynthesis propagate throughout most life.
- Further back, ribosomal RNA, or the sugars and polymers of basic cell structure exist as a deeper homology, and exist as sequences throughout most life.
So,
- The serial model best applies to newly evolved genes, alterations of allele frequencies in small populations, or alteration to genome DNA of host individuals each generation in a traceable lineage.
- The radial model best applies to matured genes that have already swept to fixation, and are widely distributed throughout populations, species, classes or even phyla.
Still, all current models of gene flow are serial. Yet, the serial model does not explain many large-scale effects of gene flow such as increase of species complexity, evolution of new classes or phyla, sexual reproduction, or concerted evolution.

5.3 Gene "Selfishness"
Any theory of gene propagation must account for gene drive, or "selfishness". Nobody proposes that genes have intent in a moral sense, but DNA sequences do tend to self-perpetuate, so we must explain why.
- We presume that life evolved from a need of order to self-perpetuate. This leads to a theory that phenotype life exists to self-perpetuate the original sequences. (The original "selfish gene" concept.)
- Only while DNA self-perpetuates in modern life, in early life DNA might not yet have evolved. Plus even in modern life genomes are so complex that small segments of DNA might not be individually affect selection. (Selection is of entire phenotypes.) Moreover, selected DNA tends to hold sequence better than non-selected DNA, which shows much of gene selfishness as a phenotype effect after all.
- Yet there is also "selfish" (or "ultra-selfish") parasitic DNA, which avoids exposure to phenotype selection, but will stealth or hitchhike its propagation via fit hosts. (Like a virus, it self-perpetuates entirely in its own interests. This is not quite the "selfish gene" concept of Richard Dawkins, but it demonstrates that DNA does self-replicate.)
A more logical explanation of gene drive is that evolution is more efficient (and hosts benefit) if any already stable DNA sequence tends to be conserved. This begins in early life. Early mutation provided evolutionary variety (a benefit) but also lethal disruption (harm) to hosts. So, mutation rate evolved (or the rate was high but repair machinery evolved) to select an optimum balance between variety and disruption. Only there is a way to optimize this further.
- If selection kept some sequences stable for a long time, they were probably useful, so it would pay to further conserve these.
- This would allow increase in overall mutation rate (to allow more variety) without risking dangerous mutations to crucial, established sequences. (Or if mutation rate was high, DNA repair machinery optimized to conserve the stable sequences.)
- Early organisms that better preserved useful sequences were able to vary type (adapt) faster than rivals, while preventing lethal disruption to crucial, well-established sequences.
In modern life too it will benefit hosts to conserve established sequences.
- Genes expressing basic materials and body plans took huge evolutionary efforts to perfect, so altering them is more likely to produce lethal disruptions than an improvement.
- But if variety is needed sequences that have altered often might better tolerate change.
- The algorithm could be simple. If a sequence had persisted for long time, the DNA repair machinery would be selected to conserve it. But if sequences had changed often there might be a less priority on repairing them.
So, gene drive reflects evolutionary efficiency at the DNA level, which tends to conserve established sequences. Over the process of life it becomes easier for large organisms to adapt (increasing evolvability). As organisms become complex large changes of morphology are possible from evolution of fewer new genes. Genes (like hox genes) become optimized at expressing complex attributes. This offers a fitness advantage to genes, because the same set of genes can express a huge variety of types. Again, we see this gene distribution pattern in;
- Placental mammals, where 70-90% of genes in common can express an extensive variety of mammal types.
- The most developed mammals, the primates, where 90-98% of genes can express a variety of rapidly adapting and fast evolving types.
- The most recently evolved large mammals, humans. Here the modern genome barely needs to alter, because the gene set expresses such an adaptable type that most adaptation can be concentrated outside of biology, in behavior and culture. (Humans have less genetic variety than chimps, though this might have another explanation, such as an early 'bottleneck' effect.)
However, if genes were truly "selfish" the best strategy for perpetuating sequence would be to express homologous attributes useful in a variety of species. These genes could retain original sequence among many types, but "force" variability onto other, later evolving genes so that other would accept the burden of change.

5.3 The Gene Spectrum
Einstein said that theories should be as simple as possible -but not simpler. The concept that DNA will try to conserve sequence is simple. Yet, as genomes composed of DNA evolve greater complexity they lose sequence against simpler genomes, from which they evolved. Critics dispute this, but eukaryote genomes with non-selected, non-nuclear and extra-genetic DNA loose sequence over simpler prokaryote genomes from which they evolved. Plus genomes in sexually reproducing species can lose 50% sequence every replication against asexual genomes, from which they evolved.
So, although we want to keep the theory simple, we must also explain why conserving sequence among small amounts of crucial, earlier-evolving DNA can force a loss of sequence on the later-evolving DNA, that makes up complex genomes.
The effect can be explained if we allow that strategies of gene propagation will form a spectrum of possibilities. If genes truly seek to replicate exact copy among the broadest variety of types, there will be one function, most likely RNA or an enzyme, that does this best. That place is then occupied in the spectrum. H4 has the highest copy fidelity in eukaryote life so it occupies that place, but there are other histones, like H1 or H2, that occupy the next best places. Histones are just an example. Life had evolved for two billion years prior to histones so the prime choices were taken-up long before that. Just as life builds in layers from simple functioning cells, which evolve first, up to more complex types which evolved from the simple ones. Early genes are the most stable occupying the choice places at the primal end of the spectrum. While at the other end, where nothing is fixed, the spectrum vibrates erratically where modern genes and sequences jostle around, trying to occupy places occupied by more stable genes.
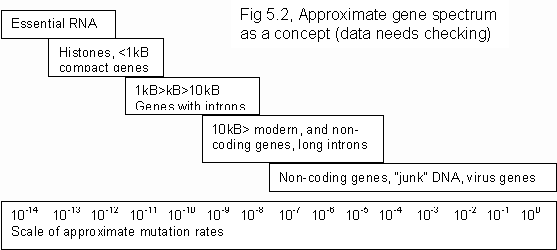
Figure 5.2 shows an approximate gene spectrum. Note that:
- Genes expressing the homologous traits of life tended to evolve earlier, and so are shorter, more prokaryote-like and harder to alter. While genes expressing easily varied homoplastic traits tend to be more recently evolved, longer eukaryote-like compound genes, which do tend to be easier to alter.
- As life evolves the DNA of modern organisms becomes longer and easier to change, but crucial sequences of life become highly matured and stabilized.
- As complex organisms increase adaptability, evolutionary cycle times go down. It becomes even easier for early evolved, long established genes to move outside the "residence" of processes that might alter them.
- In adaptable organisms the varieties of gene expression go up through sexual redistribution of alleles, enhanced expression techniques, and polygenic variation of traits. But as evolutionary variety is achieved by sophisticated means (the evolution of evolvability) need for alteration of established genes, or evolution of totally new ones goes down.
- If genes are truly "selfish" they will try to occupy the prime (rightmost) positions within the spectrum.
This spectrum effect arises because evolution has proceeded for a long time, and because some properties of life (and the universe) are easier to alter than others. In early life cells must have been close to minimum complexity, but some attributes would still have been easier to change than others. The material of a cell wall is harder to alter than cell diameter. Yet, because it is hard-to-alter but all cells need a wall, different size and shape of cells require sequences expressing cell wall proteins or other basic functions such as replication machinery. So the more different designs of cells that evolve the more widely distributed and less exposed to random extinction genes expressing cell homology will be.
Complex life is more delicate than simple life, so for this and other reasons complex life is less fit. But complexity always builds by extending the spectrum rightwards by find new gene and DNA types to express the additional complexity. The recently evolved genes are less fit (hold sequence with less fidelity over time) but at least they get to exist, plus they extend the radiation of the earlier evolved types.
We will call genes that retain sequence as their host genomes alter "smart" because such genes play the fitness odds so that they always win. While individual genes in a lineage can perish with an unsuccessful host, the gene sequence avoids genetic death by living on in other lines, or other species, classes or phyla. So, when individuals fail to reproduce, or if a species is wiped out, unique genes, DNA, and unique allele arrangements and distribution frequencies are lost. But sequences of crucial long-established genes are not lost or altered (suffer "genetic death") regardless of the fitness loss of huge numbers of individual hosts.
The prime example of core gene sets preserving sequence against a loss of host genome fitness is sexual reproduction.
- Rearrangement of alleles is an efficient way to express genotype variety, because it allows variety every gamete without requiring gene mutation, at least for short, compact genes. (Genomes of sexually reproducing organisms do accumulate longer, complex genes, more prone to mutation during sexual exchange.)
- But sexual reproduction incurs a loss of host genotype fitness. (The unique pattern of non-expressed DNA can suffer a 50% loss of sequence each gamete and unique alleles may not be passed on.)
- Yet each gamete genes expressing homologous traits will be 100% transmitted, and they gain fitness from sexual reproduction. This is because sexual rearrangement provides variety, and the more varied types that exist, the greater the chance that for any event (such as extinction) some types will be varied enough to survive.
- Interestingly, DNA that loses fitness each gamete (mostly non-expressed sequences) evolved after sex began. (In non-sexual prokaryote life, most DNA is expressed.) So while sexually reproducing individuals only pass on 50% of their DNA each gamete, we need to measure how many of original genes (like Histones) present when sex evolved, do not pass 100% of sequence each gamete.
The riddle of sex is that early genes gained fitness from it, but later evolving genes, allele arrangements and unexpressed DNA (which mostly evolved later) was forced to bare the burden of greater change. One advantage of sex was increasing expressed variety each gamete at less demand for genes to mutate for equivalent variety. This would allow genes to 'slow down' mutation rate. (Sex provides better ways to obtain variety.) This will increase fitness of the core genes, but not in the way that we would measure for a total genome that bore the burden of a 50% fitness loss in order that a privileged set of genes could enjoy a fitness gain.
Note 1: The concept that sex arose when some genes "tricked" others into accepting the burden of change has been hinted by Dawkins. But the mechanism of how this was implemented, especially in the extra steps of cell division etc. is still not known.
Note 2: Some eukaryote organisms can reproduce asexually, but most of them do not. (So if some can, why do they not all, if it is better fitness?) In the new theory it does not matter. Whether sexual or asexual, the conserved sequences will not suffer sequence loss either way.
These trends of evolving complexity as a spectrum of propagation strategies of genes set in ancient life carries into modern life. Evolution tends towards a state (seen in the great ape genome) where most genes are orthologous within a variety of types. Plus the algorithm might be simple. (If a combination works retain it, but if the combination has not stabilized, keep altering it until it does.) We do not know how these rules work at the molecular level, or if they work. Just that useful genes gain fitness by such rules, although other genes and DNA would be forced into loss of fitness (frequent alteration) to benefit the stable sequences.

5.4 Phase Shifts
In the history of a life of a gene, genes transform from a model of serial propagation (Fig 5.1a) to radial propagation (Fig 5.1b). In the new theory we call this change a "phase shift".
The "phase shift" is simply an effect. Roughly, a gene expresses and essential homology that radiates into a variety of types. Following that, individuals who fail to select the crucial sequence do not reproduce. We might say that the new, recently evolved types are more delicate, in that they cannot survive without possessing the more complex homologous attributes. But the effect on the gene is a "slowing down" in its mutation rate.
- H4 now mutates at about 10-12. But there was a time in the history of life when this sequence either did not exist, or it existed in an earlier form evolving at a faster rate. Once it "fixed" as an essential homology, it was then held to a higher conservation rate by either host or DNA-level selection, or a combination.
- Although H4 is exceptionally conserved, we infer that all genes evolved a similar way. All genes either;
- Did not exist once, so they macro-mutated into existence as a 100 step change.
- The gene was earlier evolving at an average (about 10-5) rate and became "fixed" as a new homology, and then stabilized to a slower rate.
Mathematically a "phase shift" is (by human convention) a three to four "part" slow down, as one would get from a 10-5 to 10-8 rate (slower by three "parts"). Yet the "average" mutation rate varies only two parts (from 10-5 to 10-7), which implies that no genes "phase shift". But all genes that now mutate slowly must have altered faster in the past, even at the 100 rate of a step change the first time they appeared. So, at some point the rate did slow by 3 to 4 parts. Plus the rate slow down will correlate to how genes evolved.
- The average 10-5 to 10-7 rate will be for complex, later evolving genes. H4 mutates at 10-12, but many shorter (less than 104) bp genes expressing basic homologies (which evolved early and resemble prokaryote genes) mutate at 108 to 109.
- The mutation rate will correlate with the broadness of its homology. Species can evolve in about 105 generations, but a family might take 106, an order 107, a class 108, a phylum 109 or kingdom above 1010 generations and so on (roughly). So a gene that evolves to stability within a species (it evolves in 105 or less generations) will "phase shift" if attributes of that species become the founding homology of a genus, family, class or greater.
Still, critics protest that a mutation rate is only a probability. A 10-5 mutation rate does not mean that if species evolves in 105 generations one gene will evolve exactly one new mutation in that time.
- Sharks, lungfish, mollusks, and other stable species have residences far greater than 105. (Cyanobacteria must have residence above 1010). Depending on population size genes among these species would mutate thousands of times within residence, but no new genes or new species emerged.
- Only when species evolve, it is usually among small, isolated, rapidly changing populations. Mutations can occur any time as a product of population and mutation rate. (A 10-5 mutating gene in a 109 population produces 104 mutations every generation. But it is problematic if any of these will "fix" in such a large population.
Every generation a species (like humans) evolves racially through redistribution of allele frequencies and polygenic traits. But totally new genes will only evolve when a mutation;
- is enhancing rather than disruptive
- is fit for the prevailing conditions of struggle
- 'sweeps' to fixation within a given population.
Genes mutating much slower than 10-6 typically will not have the opportunity to do this among modern species. They might have had the opportunity in the past when species evolved at slower rates (106 to 108), though there will be exceptions to every case. For example, eukaryote life must have first evolved as a single species or narrow group, so genes (like H4) expressing the homologies of eukaryote life evolved as part of the founding species. As the type stabilized and radiated, the founding genes "phase shifted" by slowing mutation to a rate expressing the broadness of the new homology.
- Once H4 slows to 10-12 its mutation rate it moves outside the "residence" of all species that are likely to alter it.
- But the way that H4 could move outside of the "mutation window" of species likely to alter it will apply for any genes. None will be able to move as far as H4, but none need to.
- To escape mutation in modern species, genes would only need move outside a 10-5 window, which correlates with modern rates. DNA point mutates about 1 bp each 109 gametes. (The range is broader, so we must explain why the range varies.). This point rate would produce a mutation in 105 generations only for genes longer than 103 to 104 bp's (it depends on many factors). Yet, these longer, more complex genes are likely to be more recently evolved and less likely to express homologies evolved early in life.
- Alteration in genes longer than 104 bp will more likely affect speciation within a family or genus. But evolution of new classes or phyla takes about 107 to 109 generations. This period allows alteration of shorter, less complex, earlier evolved genes, and those with slower mutation rates. (There will be recently evolved short genes, but these are likely to mutate faster, with other exceptions to each case.)
- Only because of phylogenic saturation, evolution of new classes and phyla is now unlikely barring a major extinction. So if an evolving gene was mutating at 10-5 and it "phase shifted" to a 10-6 to 10-8 rate, it would move "outside" a window of further significant mutation of its sequence for present life on Earth.
Still, a "phase shift" is a measurement, not a process. Except the only measurement we now take of gene propagation is host fitness (serial propagation). For small-scale evolution this measurement is sufficient, but as we saw with the evolution of sex this measurement is unrepresentative of the underlying process. (It leads to anomalies, such as a novelty evolving but host fitness appears to fall.) So, we need two ways to measure gene propagation depending on the gene type.
- We measure gene fitness as host fitness for alleles unique to any host. This explains small-scale, microevolution.
- For large-scale changes of life, we must summate gene fitness as tiny increases in host fitness over millions of generations. We measure a "phase shift" advantage that genes could undergo by expressing new homologous traits useful in a broad range of species.
Only in the past, more phase shifts were possible, because not all the major homologies of life had evolved.
- During the age of reptiles, live birth, body fur, four-chamber heart, and feathers for birds had a chance to evolve fresh homologies. During this period, genes expressing these traits 'phase-shifted' from expressing the evolving homoplastic traits at a 10-5 rate (approximately) to expressing new homologies at a rate above 10-6 (say).
- While new bird species could evolve in 104 generations by altering shape, color, behavior or diet the homology "feathers" will not change in 108 generations. (This needs to be checked. What are the rates of change of sequences expressing "feathers", as against say, wing shape or other morphology of birds?)
Only the more life advances, the harder it becomes to express fresh homologies. (Because of phylogenic saturation.) But traits that increase variability usually evolve into homologies, even if minor ones for an order or family. (Once evolution acquires means to adapt new types it usually will.) Thus,
- Genes expressing attributes of the ancestral great apes evolved minor homologies, because from a base 96% shared gene set it is possible to rapidly evolve several varied types in short evolutionary times.
- In human evolution, a few percent more of great ape genes also found a way to "phase shift" towards expressing new homologous traits (large brain, etc.). This also led to radiation and high adaptability of type, even if this time confined within a single biological species.
In summary;
- Because current theory calculates allele distribution as a function of host fitness, or host genome fitness as a function of host phenotype fitness is does not meant that all genes within hosts equally depend on host fitness. Total DNA and its unique allele distribution depend on the host for fitness, but genes expressing homologous traits of life will be less than 3% of the total host DNA in modern eukaryote genomes.
- This core of genes will be widely distributed, not only in individuals within a species, but depending on the evolutionary history of the gene, throughout families, classes, or even phyla.
- This small, privileged core of genes does not propagate serially from host to host the way host DNA fingerprints or unique allele arrangements do, despite that every gene or DNA string replicates within hosts to exist. Instead, such genes radiate through a variety of hosts, physically within hosts but "out of phase" with unique, individual host interests.
- The history of genes is to evolve within hosts, but as successful lines become the founding species of new homologies, genes radiate with the successful traits they express.
- We can measure this radiation as a "phase shift" in the mutability of the gene, as it stabilizes from a serially evolving, altering gene, to a radiating, homologous, stable one.
But though the process looks complex, it is only because the model of evolution itself is too simplified. We have a certain model we use to calculate how genes redistribute their frequency in populations, or host phenotype fitness expresses its genome fitness. But these are events occurring once per generation. Only evolution of totally new genes, or a "phase shift" from an altering gene into a stable one is an event occurring in hundreds of thousands or millions of generations. So, for a complete picture of evolution we need to show both processes.

Return to
A New Model of Evolution Home Page
Contact Information


